Enzyme
|
|
Neuraminidase_Ribbon_Diagram.jpg
An enzyme (in Greek en = in and zyme = leaven) is a protein, or protein complex, that catalyzes a chemical reaction and also controls the 3D orientation of the catalyzed substrates. Like any catalyst, enzymes work by lowering the activation energy of a reaction, thus allowing the reaction to proceed to its steady state or completion much faster than it otherwise would; the enzyme (again, as with any catalyst) remains unaltered by the completed reaction and can therefore continue catalysis. It is important to note that, as with all catalysts, all reactions catalyzed by enzymes must be 'spontaneous' (containing a net negative Gibbs free energy), i.e. with the enzyme, they run in the same direction as they would without the enzyme, just more quickly; the concept is similar to the likelihood of a ball rolling down a hill versus the likelihood of it rolling up the hill. Catalysis by an enzyme is analogous to removing a pebble that is stopping the ball from rolling down the hill; the reaction goes to completion more quickly, but the final product is identical. Given a particular starting set of conditions, the end products of a particular reaction (including net energy), once steady state is reached, must always be identical, independent of the specific individual pathway taken from beginning point to end point. This is required by the Law of Conservation of Energy, which would be violated by the possibility of a cycle of moving down a pathway releasing less net energy and back up a different pathway with higher net energy, or vice versa. An enzyme can, however, run a normally non-spontaneous reaction 'backwards' by coupling it to a spontaneous one, as long as the net free energy from the total of both reactions is negative.
Enzymes are necessary within biological cells because most chemical reactions required by the cell would occur too slowly to sustain life; oxidation of organic food compounds to provide energy is one example. Enzymes may speed up biochemical reactions by a factor of many thousand times. When, and even whether, a particular reaction will take place in a cell is governed by the presence/absence of an appropriate enzyme catalyst, and also by modulation of the activity of the enzyme.
| Contents |
Etymology and history
The word enzyme comes from Greek: "in leaven". As early as the late 1700s and early 1800s the digestion of meat by stomach secretions and the conversion of starch to sugars by plant extracts and saliva were observed.
Studying the fermentation of sugar to alcohol by yeast, Louis Pasteur came to the conclusion that this fermentation was catalyzed by "ferments" in the yeast. In 1897, yeast extracts were used to ferment sugar independent from the living yeast cells by Eduard Buchner, proving that the fermentation was caused by molecules that functioned even outside the cells. He called these molecules enzymes.
Structure and function
Activation2.JPG
In enzymes, as with other proteins, function is determined by structure. An enzyme can be a monomeric protein made up of about hundred amino acids or more, or an oligomeric protein consisting of several monomers, different or identical, that act together as a unit. As with any protein, each monomer is actually produced as a long, linear chain of amino acids, which folds in a particular fashion to produce a three-dimensional product. While it is well established that a protein's fold is determined by its primary structure, the factors that go into ensuring that the chain folds correctly and maintains its shape are complex, and still not completely understood, although general principles have been identified. Individual monomers may then combine via non-covalent interactions to form a multimeric protein. The final enzyme molecule might also be modified in various ways to make it active; for instance, chymotrypsin, a digestive protease, is produced in inactive form as chymotrypsinogen in the pancreas and transported in this form to the stomach where it is activated. This prevents the enzyme from harmful digestion of the pancreas or other tissue. This type of inactive precursor to an enzyme is known as a zymogen.
An enzyme contains one or more binding sites where the substrate(s) attach, and active site(s), where the amino acids perform the catalysis; and frequently one or more other binding sites that serve regulatory functions, which increase or inhibit the enzyme's activity. These typically bind metals or small molecules, often direct or indirect products or substrates of the reaction catalyzed. This provides a means for feedback to keep the products and substrates in proper relative abundance.
As a consequence of this basic structure, the amino acid chain of an enzyme tends to consist of one or more active sites, separated by stretches whose purpose is mainly to position the active regions correctly. Because the precise structure of each region tends to be fairly critical to correct function, and because the frequency of a mutation which would produce a nonfunctional active region is proportional to the length of the chain separating the amino acids involved, evolution works against having the amino acids from an active region widely dispersed, instead tending to keep the amino acids involved in each active region compacted fairly closely together in the chain and conserved against mutation, separating these regions by long stretches of 'spacer' amino acids where mutation is much less critical (although some mutations in these regions can also inactivate the product). This has the additional effect of making each region act, relative to mutation, somewhat like an independent subunit which can be duplicated, deleted, moved, or 'mixed-and-matched' with other such regions, generating new proteins to be tested for evolutionary success. This would seem to be a more efficient process in terms of the likelihood of successfully generating functional new enzymes than would having the functionally related amino acids of the active site widely dispersed throughout the amino acid chain, with random mutation occurring anywhere.
Factor_D_Enzyme.jpg
Enzymes can couple two or more reactions, so that a thermodynamically favourable reaction can be used to "drive" a thermodynamically unfavorable one, in effect running the reaction 'backwards', or 'uphill'. For instance, the high energy compound ATP is generated in the cell by coupling its synthesis to the oxidation of sugars, which releases more energy than the syntehsis of ATP requires; then the ATP is broken down in turn by other enzymes coupled to other processes, releasing the energy stored in it in order to drive other, otherwise energetically unfavorable, chemical reactions. This is analogous to raising a heavy object by attaching it to a rope that goes over a pulley and attaches to a weight which is lowered; the second weight must be heavier, in order that the net energy balance of the sum of both actions is favorable (i.e., net energy in the system is lowered).
In a very similar fashion, an enzyme can cause an easily reversible reaction to become essentially irreversible, by coupling it to another, energetically unbalanced reaction. As with any catalyst, an enzymatic pathway does not intrinsically change the reversibility or irreversibility of a reaction. The direction of any chemical reaction is based only on the energy balance between the two sides of the reaction equation, and if there is very little change in net free energy between the substrates and the products, the reaction will normally be easily reversible, running in either direction depending on which side of the reaction was more highly concentrated. However, enzymes provide a means to make such a reaction much less reversible, by coupling the reaction to one with a large release of energy. This is analogous to an object sliding freely on a flat surface; the object may be pulled in one direction by attaching it to a rope attached to a hanging weight. In order to lower the total net energy, the system will drive the coupled reactions in one direction.
Rate of enzyme mediated reactions
Main article: Rate of enzyme mediated reactions
Enzymes can increase reaction rate by favoring or enabling a different reaction pathway with a lower activation energy, making it easier for the reaction to occur. The overall rate of enzyme mediated reactions depends on many factors.
Specificity
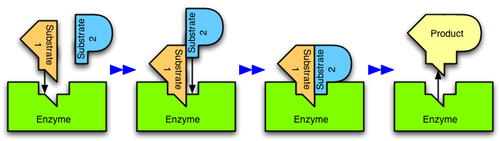
Enzymes are usually specific as to the reactions they catalyze and the substrates that are involved in these reactions. Complementary structural properties of the enzyme and substrate are responsible for this specificity (Fig. 2).
Kinetic
In 1913, Leonor Michaelis and Maud Menten proposed a quantitative theory of enzyme kinetics which is still widely used today (usually referred to as Michaelis-Menten kinetics). Enzymes can perform up to several million catalytic reactions per second; to determine the maximum speed of an enzymatic reaction, the substrate concentration is increased until a constant rate of product formation is achieved. This is the maximum velocity (Vmax) of the enzyme. In this state, all enzyme active sites are saturated with substrate. However, Vmax is only one kinetic parameter that biochemists are interested in. The amount of substrate needed to achieve a given rate of reaction is also of interest. This can be expressed by the Michaelis-Menten constant (KM), which is the substrate concentration required for an enzyme to reach one half its maximum velocity. Each enzyme has a characteristic KM for a given substrate. Since Vmax cannot be measured directly, both KM and Vmax are usually determined by extrapolating from a limited data set, using what is known as a double reciprocal, or Lineweaver-Burk plot.
The efficiency of an enzyme can be expressed in terms of kcat/Km. The quantity kcat, also called the turnover number, incorporates the rate constants for all steps in the reaction, and is the product of Vmax and the total enzyme concentration. kcat/Km is a useful quantity for comparing different enzymes against each other, or the same enzyme with different substrates, because it takes both affinity and catalytic ability into consideration. The theoretical maxium for kcat/Km is about 108 to 109 (mol/L)-1s-1. At this point, every collision of the enzyme with its substrate will result in catalysis. Some enzymes, such as fumarase, actually approach this limit.
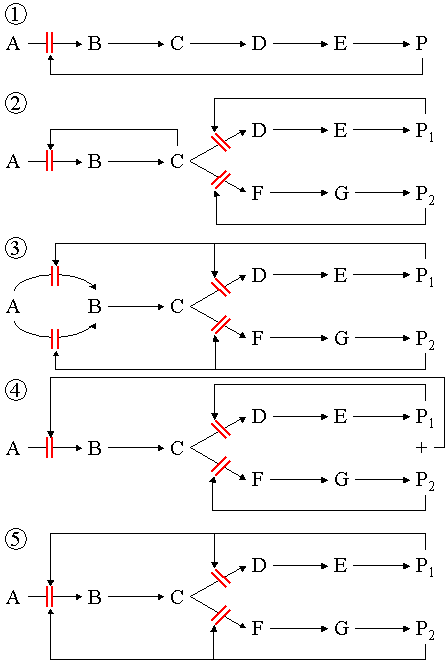
Metabolic pathways
Several enzymes can work together in a specific order, creating metabolic pathways. In a metabolic pathway, one enzyme takes the product of another enzyme as a substrate. After the catalytic reaction, the product is then passed on to another enzyme. The end product(s) of such a pathway are often inhibitors for one of the first enzymes of the pathway (usually the first irreversible step, called committed step), thus regulating the amount of end product made by the pathway.
Quantum-mechanical model of enzyme catalysis
The lecture "Quantum Theory of some Biochemical Reactions", presented to the IV International Biophysical Congress (Moscow, 1972) by R.R. Dogonadze and Z.D. Urushadze, formulated the first quantum mechanical model of the simplest form of enzyme catalysis. In 1972-1973, in the works of M.V. Volkenshtein, R.R. Dogonadze, A.K. Madumarov, Z.D. Urushadze and Yu.I. Kharkats were formulated the quantum-mechanical (physical) model of Enzyme Catalysis. These works demonstrated the role of conformational transformations in catalytic reactions.
Ribozymes
An RNA enzyme or "ribozyme" is made of RNA instead of protein. Generally, ribozymes only catalyze RNA splicing, though one notable exception is the RNA portion of the ribosome, which catalyzes the formation of peptide bonds in growing protein chains.
Enzymes and health
Enzymes are essential to living organisms, and a malfunction of even a single enzyme out of approximately 2,000 types present in our bodies can lead to severe or lethal illness. An example of a disease caused by an enzyme malfunction in humans is phenylketonuria (PKU). The enzyme phenylalanine hydroxylase, which usually converts the essential amino acid phenylalanine into tyrosine does not work, resulting in a buildup of phenylalanine that leads to mental retardation. Enzymes in the human body can also be influenced by inhibitors. Aspirin, for example, inhibits an enzyme that produces prostaglandins (inflammation messengers), thus suppressing pain and inflammation. Enzymes are also used in everyday products such as washing detergents, where they speed up chemical reactions involved in cleaning the clothes (for example, breaking down blood stains).
Digestive and metabolic enzymes
Nutrition in animals relies on digestive enzymes such as salivary amylase, trypsin and chymotrypsin. Their primary role is for the digestion of food and making nutrients available to all of the body processes which need them. Another class of enzymes is called metabolic enzymes. Their role is to catalyze chemical reactions involving every process in the body, including the participation of oxygen. Most of our cells (an exception being erythrocytes), would literally starve for oxygen even with an abundance of oxygen without the action of the enzyme, cytochrome oxidase. Enzymes are also necessary for muscle contraction and relaxation. The fact is, without both of these classes of enzymes, (digestive and metabolic) life could not exist.
Enzyme naming conventions
By common convention, an enzyme's name consists of a description of what it does, with the word ending in "-ase". Examples are alcohol dehydrogenase and DNA polymerase. Kinases are enzymes that transfer phosphate groups. The International Union of Biochemistry and Molecular Biology has developed a nomenclature for enzymes, the EC numbers; each enzyme is described by a sequence of four numbers, preceded by "EC". The first number broadly classifies the enzyme based on its mechanism:
- EC 1 Oxidoreductases: catalyze oxidation/reduction reactions
- EC 2 Transferases: transfer a functional group (e.g., a methyl or phosphate group)
- EC 3 Hydrolases: catalyze the hydrolysis of various bonds
- EC 4 Lyases: cleave various bonds by means other than hydrolysis and oxidation
- EC 5 Isomerases: catalyze isomerization changes within a single molecule
- EC 6 Ligases: join two molecules with covalent bonds
The complete nomenclature can be browsed at http://www.chem.qmul.ac.uk/iubmb/enzyme/
Enzymes and classes of enzyme
- Adenylate cyclase: participate in signal transduction by converting ATP into cyclic AMP
- Alcohol dehydrogenase: breaks down alcohols to aldehydes in human liver; creates alcohol in yeast fermentation
- Alkaline phosphatase
- Amylase: breaks starch into maltose, present in human saliva and also released by the pancreas
- Angiotensin converting enzyme
- ATPases: those enzymes that hydrolyze ATP into ADP and inorganic phosphate in order to drive some other energetically unfavorable reaction
- ATP synthase: creates ATP from ADP, inorganic phosphate and some form of energy
- Autolysin: general term for enzymes that cause cells to die
- Beta-galactosidase: See lactase.
- Beta-lactamase: breaks down penicillin, rendering those bacteria that produce beta-lactamase resistant
- Catalase: converts hydrogen peroxide to oxygen and water
- Chitinase: breaks down chitin
- Cholinesterase
- Chymosin: better known as rennet, contained in the stomachs of many animals and causes milk to curdle
- Chymotrypsin: can digest proteins
- Coenzyme Q - cytochrome c reductase: participates in the electron transfer chain by reducing cytochrome c and pumping protons across the membrane
- Cytochrome c oxidase: endpoint of the electron transfer chain, where oxygen is reduced to water
- Cytochrome c peroxidase takes reducing equivalents from cytochrome c and reduces hydrogen peroxide to water
- Deiodinase: activates thyroid hormones by the removal of iodine
- Diastase: break starch into glucose
- Dihydrofolate reductase: reduces dihydrofolic acid to tetrahydrofolic acid
- DNA gyrase: catalyzes the negative supercoiling of prokaryotic DNA
- Glutamate decarboxylase: synthesizes the neurotransmitter GABA in the brain
- Glutathione peroxidase: protects cells by converting hydrogen peroxide to water
- Isomerase: all those enzymes which convert a molecule into one of its isomeric forms
- Kinases: all those enzymes which catalyze the transfer of phosphate groups
- Lactase (or beta-galactosidase): breaks lactose into galactose and glucose
- Ligases: all those enzymes that can join ("ligate") two molecules with a covalent bond
- Lyases: all those enzymes that can eliminate a group from a molecule to form a double bond and vice versa.
- Monoamine oxidase (MAO): oxidize certain neurotransmitters and biologically active amines
- NADH dehydrogenase: the first complex in the electron transfer chain, transferring electrons from NADH to coenzyme Q
- Nitrogenase
- Ornithine decarboxylase: processes ornithine as the first step in the production of polyamines
- Peroxidase: the group of enzymes that can convert hydrogen peroxide or similar compounds
- Phenylalanine hydroxylase (PAH):
- Phospholipase
- Polymerase: builds long chains of nucleic acid polymers from their constituents
- Protease (peptidase): those enzymes which cut peptide bonds of proteins
- Protein kinases: those enzymes that transfer a phosphate group to an amino acid residue of a protein
- Protein phosphatases: those enzymes that take off a phosphate group from an amino acid residue of a protein
- Restriction enzymes: those enzymes which cut DNA or RNA at specific locations
- Reverse transcriptase: used by retroviruses to transcribe information from RNA into DNA
- Ribulose bisphosphate carboxylase: fixes carbon in green plants.
- RNAse: those enzymes which can break down RNA into the component nucleotides
- Superoxide dismutase: transforms superoxide into oxygen and hydrogen peroxide
- Trypsin: digests proteins
- Tyrosine kinases: those enzymes which transfer phosphate groups to a tyrosine residue in a protein
- Tyrosinase
- Urease: hydrolizes urea into carbon dioxide and ammonia
- Xanthine oxidase: oxidizes hypoxanthine to xanthine and further to uric acid
Purification
Since enzymes are proteins, enzyme purification begins with protein purification. Each step in the purification procedure is monitored for enzyme activity. One caveat is that proteases (enzymes that digest proteins) can be expected to start digesting themselves as they become more concentrated unless their activity is inhibited somehow; in a similar vein, purifications of other enzymes that begin with a preparation containing a protease will often result in loss of the enzyme being purified unless the protease is inhibited or inactivated.
See also
References
- Koshland D. The Enzymes, v. I, ch. 7, Acad. Press, New York, 1959
- Perutz M. Proc.Roy.Soc., B 167, 448, 1967
- M.V. Volkenshtein, R.R. Dogonadze, A.K. Madumarov, Z.D. Urushadze, Yu.I. Kharkats. Theory of Enzyme Catalysis.- Molekuliarnaya Biologia, Moscow, 6, 1972, pp. 431-439 (In Russian, English summary)
- M.V. Volkenshtein, R.R. Dogonadze, A.K. Madumarov, Z.D. Urushadze, Yu.I. Kharkats. Electronic and Conformational Interactions in Enzyme Catalysis.- In: E.L. Andronikashvili (Ed.), Konformatsionnie Izmenenia Biopolimerov v Rastvorakh, Publishing House "Nauka", Moscow, 1973, pp. 153-157 (In Russian, English summary)
- R.R. Dogonadze, Z.D. Urushadze, V.K. Khidureli. Calculation of Kinetic Parameters of Reactions with the Conformational Transformations.- In: E.L. Andronikashvili (Ed.), Konformatsionnie Izmenenia Biopolimerov v Rastvorakh, Publishing House "Metsniereba", Tbilisi, 1975, pp. 368-375 (In Russian)
External links
- ExPASy enzyme database (http://us.expasy.org/enzyme/), links to Swiss-Prot sequence data, entries in other databases and to related literature searches
- PDBsum (http://www.biochem.ucl.ac.uk/bsm/enzymes/) links to the known 3-D structure data of enzymes in the Protein Data Bank
- BRENDA (http://www.brenda.uni-koeln.de), comprehensive compilation of information and literature references about all known enzymes; requires payment by commercial users
- Weizmann Institute's Genecards Database (http://bioinformatics.weizmann.ac.il/cards/), extensive database of protein properties and their associated genes.
- Cytochrome P450 enzymes (http://drnelson.utmem.edu/CytochromeP450.html) site lists over 4000 versions of enzymes from this cytochrome in plants and animalsbg:Ензим
cs:Enzym da:Enzym de:Enzym es:Enzima eo:Enzimo fa:زیمایه fr:Enzyme id:Enzim io:Enzimo it:Enzima he:אנזים nl:Enzym ja:酵素 pl:Enzym pt:Enzima ru:Энзим simple:Enzyme sl:Encim su:Énzim fi:Entsyymi sv:Enzym tr:Enzim zh:酶