Translation (genetics)
|
|
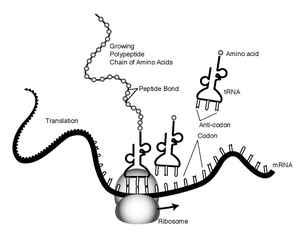
Translation is the second process of protein biosynthesis (part of the overall process of gene expression). In translation, messenger RNA is decoded to produce a specific polypeptide according to the rules specified by the genetic code. Translation is necessarily preceded by transcription. Similarly to transcription, translation proceeds in three phases: initiation, elongation and termination (all describing the growth of the amino acid chain, or polypeptide that is the product of translation). The capacity of disabling or inhibiting translation in protein biosynthesis is used by antibiotics such as: anisomycin, cycloheximide, chloramphenicol and tetracycline.
| Contents |
Basic mechanisms
The mRNA carries the genetic information from the chromosomes to the ribosomes. Harboured on the ribosome (structures containing rRNA and a protein base), the mRNA information is matched (through hydrogen bonds) to the specific tRNA (tRNA is a small RNA chain (74-93 nucleotides) that transfers a specific amino acid to a growing polypeptide chain at the ribosomal site of protein synthesis). The information sequence is tranferred in triplets of nucleotides. Each of those triplets is linked to an amino acid. Each three mRNA nucleotides are called a codon and their three complementary tRNA nucleotides are called its anti-codon. Aminoacyl tRNA synthetase is the enzyme that catalyzes the binding of a specific amino acid to a tRNA to form an aminoacyl-tRNA.
Prokaryotic translation
Prokaryotes have no nucleus, so mRNA can be translated while it is still being transcribed. The translation is said to be polyribosomal when there is more than one active ribosome.
Initiation
Initiation of translation involves the small ribosomal subunit binding to the 'start' codon on the mRNA, which indicates where the mRNA starts coding for the protein. This codon is most commonly an AUG, but alternative start codons are common in prokaryotes. In bacteria, the protein starts instead with the modified amino acid N-formyl methionine (f-Met). In f-Met, the amino group has been blocked by a formyl group to form an amide, so this amino group can not form a peptide bond. This is not a problem because the f-Met is at the amino terminus of the protein. In prokaryotes the binding of the small subunit to the correct place on the mRNA is facilitated by base pairing to a series of bases known as the Shine-Dalgarno sequence, located 8-13 nucleotides before the start site.
Elongation
The large subunit forms a complex with the small subunit, and elongation proceeds. A new activated tRNA enters the A site of the ribosome and base pairs with the mRNA. The enzyme peptidyl transferase forms a peptide bond between the adjacent amino acids. As this happens, the amino acid on the P site leaves its tRNA and joins the tRNA at the A site. The ribosome them moves in relation to the mRNA shifting the tRNA at the A site on to the P whilst releasing the empty tRNA, this process is known as translocation.
Termination
This procedure repeats until the ribosome encounters one of three possible stop codons, where translation is terminated. This stalls protein growth, and release factors, proteins which mimic tRNA, enter the A site and release the protein in to the cytoplasm.
Eukaryotic translation
In eukaryotes, transcription occurs in the nucleus, then the mRNA moves to the cytoplasm for the translation to occur. The mRNA is spliced with 5' cap and 3' poly-A-tail and then transported. Initiation is described below, elongation and termination proceed similarly to that in prokaryotes.
Initiation
Initiation of translation involves the small ribosomal subunit binding to the 'start' codon on the mRNA, which indicates where the mRNA starts coding for the protein. In eukaryotes and archaea, the amino acid encoded by the start codon is methionine. The initiator tRNA charged with Met forms part of the ribosomal complex.
Translation by hand
It is also possible to translate either by hand (for short sequences) or by computer (after first programming one appropriately), this allows biologists and chemists to draw out the chemical structure of the encoded protein on paper.
First, convert each DNA base to its RNA complement:
DNA -> RNA A -> U T -> A G -> C C -> G
Then split into triplets, and see:Genetic code for the code table used by ribosomes. Note that there are 3 translation "windows" depending on where you start reading the code. Finally, use the table at Amino acid to translate the above into a structural formula as used in chemistry.
This will give you the primary structure of the protein. However, proteins tend to fold, depending in part on hydrophilic and hydrophobic segments along the chain. Secondary structure can often still be guessed at, but the proper tertiary structure is often very hard to determine, though chemical simulations currently are able to guess right about 70% of the time.cs:Translace de:Translation (Biologie) it:Sintesi proteica ja:翻訳 (生物学) pl:Translacja fr:Synthèse_des_protéines
