Hidden Markov model
|
|
A hidden Markov model (HMM) is a statistical model where the system being modelled is assumed to be a Markov process with unknown parameters, and the challenge is to determine the hidden parameters, from the observable parameters, based on this assumption. The extracted model parameters can then be used to perform further analysis, for example for pattern recognition applications.
In a regular Markov model, the state is directly visible to the observer, and therefore the state transition probabilities are the only parameters. A hidden Markov model adds outputs: each state has a probability distribution over the possible output tokens. Therefore, looking at a sequence of tokens generated by an HMM does not directly indicate the sequence of states.
| Contents |
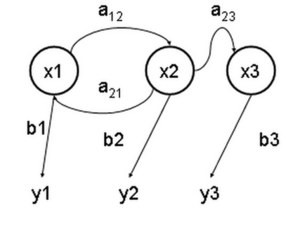
State transitions in a hidden Markov model
Evolution of a Markov model
The preceding diagram emphasizes the state transitions of a hidden Markov model. It is also useful to explicitly represent the evolution of the model over time, with the states at different times t1 and t2 represented by different variables, x(t1) and x(t2).
Hmm_temporal_bayesian_net.png
Temporal evolution of a hidden Markov model
In this diagram, it is understood that the time slices (x(t), y(t)) extend to previous and following times as needed. Typically the earliest slice is at time t=0 or time t=1.
Using Markov models
There are 3 canonical problems to solve with HMMs:
- Given the model parameters, compute the probability of a particular output sequence. Solved by the forward algorithm.
- Given the model parameters, find the most likely sequence of (hidden) states which could have generated a given output sequence. Solved by the Viterbi algorithm.
- Given an output sequence, find the most likely set of state transition and output probabilities. Solved by the Baum-Welch algorithm.
A concrete example
This example is further elaborated in Viterbi algorithm page.
Applications of hidden Markov models
- speech recognition or optical character recognition
- natural language processing
- bioinformatics and genomics
- prediction of protein-coding regions in genome sequences
- modelling families of related DNA or protein sequences
- prediction of secondary structure elements from protein primary sequences
- and many more...
See also
References
- Lawrence Rabiner, 1989. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. http://www.caip.rutgers.edu/~lrr/Reprints/tutorial%20on%20hmm%20and%20applications.pdf
- Kristie Seymore, Andrew McCallum, and Roni Rosenfeld. Learning Hidden Markov Model Structure for Information Extraction. AAAI 99 Workshop on Machine Learning for Information Extraction, 1999. (also at CiteSeer: [1] (http://citeseer.nj.nec.com/seymore99learning.html))
External links
- Hidden Markov Model (HMM) Toolbox for Matlab (http://www.ai.mit.edu/~murphyk/Software/HMM/hmm.html) (by Kevin Murphy)
- Hidden Markov Models (http://www.cs.brown.edu/research/ai/dynamics/tutorial/Documents/HiddenMarkovModels.html) (an exposition using basic mathematics)
- GHMM Library (http://www.ghmm.org) (home page of the GHMM Library project)
- A step-by-step tutorial on HMMs (http://www.comp.leeds.ac.uk/roger/HiddenMarkovModels/html_dev/main.html) (University of Leeds)de:Verborgenes Markow-Modell